Research Article
Screening and Transcriptome Analysis of Different Materials with Low Temperature Tolerance in Eggplant (Solanum melongena) 
 Author
Author  Correspondence author
Correspondence author
International Journal of Horticulture, 2022, Vol. 12, No. 4 doi: 10.5376/ijh.2022.12.0004
Received: 19 Oct., 2022 Accepted: 24 Oct., 2022 Published: 31 Oct., 2022
Zhu Z.W., Wu X.X., Zhang A.D., and Zha D.S., 2022, Screening and transcriptome analysis of different materials with low temperature tolerance in eggplant (Solanum melongena), International Journal of Horticulture, 12(4): 1-10 (doi: 10.5376/ijh.2022.12.0004)
The growth and fruit quality of eggplant were seriously affected by low temperature stress. In order to screen low temperature tolerance germplasm of eggplant and reveal its correlation with low temperature tolerance at molecular level. In this study, two eggplant inbred lines with different genotypes were selected on the basis of preliminary experiments. The effects of low temperature stress on the seed germination and seedling chilling injury were studied. The transcriptome of seedling leaves under 4°C low temperature was sequenced, and the differentially expressed genes were classified and enriched. The results showed that CHEN18 was much stronger than 819 in seed germination and seedling low temperature tolerance. The analysis of transcriptome data showed that there were some differences in genetic background between the two eggplant materials, and the differentially expressed genes were mainly concentrated in biological regulation, cell process, metabolic process and single organism process. Up-regulated differentially expressed genes are mainly enriched in cell processes, environmental information processing, genetic information processing and metabolism. The results of this study laid a foundation for the selection of cold tolerance germplasm and the excavation of cold tolerance genes in eggplant.
Eggplant (Solanum melongena L.) is a thermophilic vegetable with a long cultivation history in China. The varieties planted in different provinces are quite different, forming a characteristic of relatively obvious regional distribution (Lian et al., 2017). Low temperature stress will have a serious impact on its yield and quality (Niño-Medina et al., 2017). How to plant and market successfully under low temperature has become the goal pursued by many growers. China is rich in eggplant germplasm resources. Due to its own characteristics and the results of long-term natural selection, the low temperature tolerance is generally poor. However, there are still some wild relatives and endemic varieties in some areas, providing the possibility to improve its genetic resources (Lian et al., 2006). At present, the research on eggplant low temperature is mostly limited to the improvement of cultivation and management measures and the research on physiological and biochemical basis (Zhang and Xu, 2009; Bao et al., 2013; Wu et al., 2017; Zhang et al., 2020), which cannot fundamentally solve the actual situation of current production. In recent years, with the deepening of plant breeding for stress resistance, more and more cold tolerant germplasm screening and identification of related gene expression have received extensive attention (Dhawan et al., 2016; Ebrahimi et al., 2017; Bai et al., 2019; Wang et al., 2020). How to improve the low temperature tolerance of eggplant varieties, and how to understand the mechanism of its response to low temperature stress from the genetic material has become an important research approach.
On the basis of previous work (Zhu et al., 2019), two eggplant materials CHEN18 and 819 with significant difference in low temperature tolerance were selected for identification of low temperature seed vitality and seedling chilling injury. A simple and rapid method for identifying eggplant germplasm with low temperature tolerance was obtained by comparing seed germination and seedling tolerance to low temperature. The seedlings of the two materials were subjected to transcriptome analysis after 4°C low temperature treatment, and the sequencing quality was determined by comparing the reference genome. The correlation analysis of the detected differentially expressed genes showed that the two eggplant materials had significantly different genetic backgrounds, and a large number of differentially expressed genes were produced by low temperature treatment compared with the control. The comparison between CHEN18 treated with low temperature and 819 treated with low temperature showed that the number of differential protein coding genes is huge, which is far beyond the difference in their own genetic background.
1 Results and Analysis
1.1 Effect of low temperature on seed germination of two eggplant materials
Eggplant seed germination has certain requirements on temperature, and low temperature will seriously affect its seed germination and seed vitality. The results showed (Table 1) that the germination of CHEN18 and 0819 was affected under different low temperature treatments. At 15°C and 20°C, the seed germination rate of CHEN18 remained above 90% with high germination index and vitality index, showing a strong resistance to low temperature. However, 0819 was more sensitive to low temperature, its germination rate, germination index and vitality index were significantly lower than the control level under 20°C. At 15°C, its germination rate was lower than 10%, and at 10°C, its germination was completely limited.
|
Table 1 The effects of different low-temperature treatments on seed germination of CHEN18 and 0819 Note: The data in the table are shown as mean ± standard deviation; different lowercase letters in the same column indicate significant difference at 0.05 level |
1.2 Effects of low temperature stress on injury of eggplant seedling leaves
The chilling injury index of CHEN18 and 819 eggplant seedling leaves were observed and counted at different treatment times in a 4°C low temperature incubator. The effect of 4°C low temperature on the morphology of the two eggplant materials was very significant. The overall change range of chilling injury index of leaves after low temperature treatment of CHEN18 was small. The chilling injury index was lower than 10 during the period of 12~72 h and reached the maximum value of 17.2 after 168 h treatment. After low temperature treatment, the chilling injury index of 819 increased immediately, higher than 30 at 12 h, and reached the maximum value of 73.1 at 168 h (Figure 1).
|
Figure 1 The chilling injury index of eggplant leaves after 4℃ low-temperature treatment |
1.3 Analysis of transcriptome quality under low temperature stress
The seedlings treated at 4°C and grown at room temperature were sampled and sequenced for transcriptome respectively. Clean reads (Table 2) were obtained after removing the reads with joints, high content of unknown base N and low quality. The clean-reads of each treatment are about 50M, the quality of Q30 is more than 90%, and the GC content is about 43%.
|
Table 2 Sequence summary results of transcriptome in each sample |
HISAT2 software was used to compare clean reads with the data in eggplant genome (http://eggplant.kazusa.or.jp/). The results showed that 83.99%, 93.08%, 83.72%, and 91.39% of CK1, CK2, T1, and T2 reads are mapped to the reference genome, respectively, of which 80.50%, 89.01%, 80.61%, and 87.73% can be uniquely mapped (Table 3). The reads map to “+” and “-” of T1 and T2 after low temperature treatment on the genome are basically consistent with CK1 and CK2. The percentage of non-splice reads of T1 and T2 after low temperature treatment was 49.67% and 59.54%, which was lower than 51.21% and 61.09% of the control at room temperature. While the percentage of splice reads increased to 30.94% and 29.20%, higher than 29.23% and 27.92% of the control at room temperature.
|
Table 3 Reference genome comparison results |
The overall situation of the distribution of reads in genome region (Figure 2) showed that the percentage of reads mapped to exons is more than 80%. Although the ratio of reads map to intron and intergenic was low, it still accounted for a certain percentage, indicated that some pre-mRNA residues or variable splicing intron residues may exist in the sequencing data, and eggplant reference genome annotation needed to be further improved.
|
Figure 2 Distribution of reads in genome region in each processing Note: a: Exon; b: Intergenic; c: Introns |
1.4 Analysis of differentially expressed genes in transcriptome
To analyze the differential gene expression of two eggplant materials under low temperature treatment, we calculated the expression of protein coding genes by FPKM method and screened the differentially expressed genes with |log2 fold change|>1 and P-value<0.05 to draw the volcano-plot of differential gene expression (Figure 3). Compared with CK1, T1 produced 6 402 differentially expressed genes after low temperature treatment, of which 3 670 were up and 2 732 were down. T2 produced 4 298 differentially expressed genes after low temperature treatment compared with CK2, of which 2 177 were up and 2 121 were down. After low temperature treatment, 9 064 differentially expressed genes were produced between two different varieties. The results showed that there were significant differences in gene expression between the two eggplant materials under low temperature treatment, which provided a basis for us to select related genes for low temperature tolerance identification.
|
Figure 3 Volcano-plot of differential gene expression between two eggplant varieties treated with low temperature and control |
The GO enrichment analysis of the differentially expressed genes in the above different treatment groups showed that the differentially expressed genes between the low temperature treatment group and the control group and the low temperature treatment group of different eggplant varieties were mainly enriched in four aspects of biological regulation, cell process, metabolic process and single organism process. In the classification of cell group, it is mainly concentrated in cells, cell components and organelles. It is mainly concentrated in binding and catalytic activities in molecular functional classes (Figure 4).
|
Figure 4 Comparative distribution of differentially expressed genes and all genes at GO Level2 level Note: 1: Biological adhesion; 2: Biological regulation; 3: Cell killing; 4: Cellular component organization or biogenesis; 5: Cellular process; 6: Developmental process; 7: Establishment of localization; 8: Growth; 9: Immune system process; 10: Localization; 11: Locomotion; 12: Metabolic process; 13: Multi-organism process; 14: Multicellular organismal process; 15: Negative regulation of biological process; 16: Positive regulation of biological process; 17: Regulation of biological process; 18: Reproduction; 19: Reproductive process; 20: Response to stimulus; 21: Rhythmic process; 22: Signaling; 23: Single-organism process; 24: Cell; 25: Cell junction; 26: Cell part; 27: Extracellular matrix; 28: Extracellular matrix component; 29: Extracellular region; 30: Extracellular region part; 31: Macromolecular complex; 32: Membrane; 33: Membrane part; 34: Membrane-enclosed lumen; 35: Mitochondrion-associated adherends complex; 36: Nucleoid; 37: Organelle; 38: Organelle part; 39: Symplast; 40: Synapse; 41: Synapse part; 42: Virion; 43: Virion part; 44: D-alanyl carrier activity; 45: Antioxidant activity; 46: Binding; 47: Catalytic activity; 48: Channel regulator activity; 49: Chemoattractant activity; 50: Chemorepellent activity; 51: Electron carrier activity; 52: Enzyme regulator activity; 53: Metallochaperone activity; 54: Molecular transducer activity; 55: Morphogen activity; 56: Nucleic acid binding transcription factor activity; 57: Nutrient reservoir activity; 58: Protein binding transcription factor activity; 59: Protein tag; 60: Receptor activity; 61: Receptor regulator activity; 62: Structural molecule activity; 63: Translation regulator activity; 64: Transporter activity |
Pathway analysis of differential protein coding genes using KEGG database could help us better understand biological functions. Compared with the control, the up-regulated differentially expressed genes of the two eggplant varieties under low temperature treatment were mainly concentrated in cell process, environmental information processing, genetic information processing and metabolism, of which metabolism accounted for the largest proportion. The up-regulated differentially expressed genes of the two varieties after low temperature treatment was consistent with the above. All up-regulated expressions were mainly concentrated in glucose metabolism and signal transduction pathway (Figure 5).
|
Figure 5 Up-regulation of KEGG enrichment pathway by differentially expressed genes Note: 1~12: Metabolism; 1: Eno biotics biodegradation and metabolism; 2: Nucleotide metabolism; 3: Metabolism of terpenes and polypeptides; 4: Metabolism of other amino acids; 5: Metabolism of cofactors and vitamin; 6: Lipid metabolism; 7: Glycan biosynthesis and metabolism; 8: Global and overview maps; 9: Energy metabolism; 10: Carbohydrate metabolism; 11: Biosynthesis of other secondary metabolites; 12: Amino acid metabolism; 13~16: Genetic information processing; 13: Translation; 14: Transcription; 15: Replication and repair; 16: Folding, sorting and degradation; 17,18: Environmental information processing; 17: Signal transduction; 18: Membrane transport; 19~23: Cellular processes; 19: Transport and catabolism; 20: Cellular community-prokaryotes; 21: Cellular community-eukaryotes; 22: Cell motility; 23: Cell growth and death |
In order to compare the number of differentially expressed genes in the same variety of low temperature treatment and room temperature control groups, as well as in different varieties of treatment and room temperature control, we used TBtools to draw the Venn diagram of differentially expressed gene (Figure 6). It showed that the number of differentially expressed genes of CK1 and CK2 at room temperature was 6 902, which indicated that there were great differences in genetic background between the two varieties, providing a great theoretical basis for us to find the expression of genes related to low temperature. The number of differentially expressed genes between T1 and CK 1 was 6 402, T2 and CK2 was 4 298, and T1 and T2 was 9 064. Except for the 5 002 differentially expressed genes of the two varieties under room temperature, the number of differentially expressed genes between the two varieties was 4 062 due to low temperature treatment.
|
Figure 6 Venn diagram of differentially expressed gene |
2 Discussion
Low temperature will affect the normal metabolism of plants, which will have a serious impact on their growth (Huang et al., 2017). The osmoregulation substances, antioxidant system and enzyme activities related to metabolism in plants will change significantly under low temperature environment (Samaneh et al., 2018). These changes reflected that there was a timely response and clearing mechanism for the plant body to stress. The response of plants to low temperature stress can help us to select low temperature resistant materials to some extent. In the early stage, we conducted germination tests on eggplant seeds of different genotypes, and found that different types of eggplant seeds had different responses to low temperature. Most varieties need environmental temperature above 20°C for seed germination, while few varieties have certain germination ability under the condition of below 15°C (Zhu et al., 2019). The inbred line material CHEN18 in this experiment still has a certain germination ability when the temperature is as low as 10°C, which to some extent indicates that it has a strong resistance to low temperature or a certain degree of low temperature tolerance. As one of the most intuitive phenotypic indicators, chilling injury index has been widely used in the determination of plant low-temperature resistance (Deng et al., 2014; Bai et al., 2017; Yan et al., 2018). Through the observation of chilling injury index, we can find potential low temperature resistant materials to serve for the later identification of related genes. In the low temperature treatment experiment at seedling stage, we found that the chilling injury index of CHEN18 was far lower than 819, and the overall damage of the plant was slight, indicating its potential low temperature resistance, which is consistent with the results of cold tolerance identification of wheat varieties by Zhao et al. (2019). Cheng et al. (2012) took the chilling injury index as the most important field low temperature tolerance index in identifying cucumber core germplasm. Wu et al. (2019) successfully selected the low temperature tolerant strain of sweet pepper through the phenotypic determination of plant withering grade under the low temperature of 4°C. According to the index of seed germination and chilling injury of seedlings, we think that the eggplant material CHEN18 has strong resistance to low temperature, while 819 is a low temperature sensitive material.
In order to explain the difference between them in terms of genetic material, we sequenced the transcriptome of the two materials after low temperature treatment. As a rapidly developing molecular technology, transcriptome sequencing has been applied in more and more crops (Wang et al., 2020; Li et al., 2020). Through the analysis of transcriptome data, we can find that there are some differences in the genetic background between the two eggplant materials tested, and 83.99% and 93.08% of reads were mapped to the reference genome at room temperature, respectively. After low temperature treatment, the CHEN18 differentially expressed genes to normal temperature control was 6 402, and that of 819 to normal temperature control was 4 298. It can be seen that under the conditions of different genetic backgrounds, CHEN18 has a larger number of differentially expressed genes after low temperature treatment, and there may be key genes related to low temperature tolerance in these genes. And 9 064 differentially expressed genes were produced from two different materials after low temperature treatment, indicating that low temperature induced a series of differentially expressed genes. Sareh et al. (2020) revealed barley salt tolerance related genes mediated by rapidly triggered ion transporters through transcriptome analysis. Specific transcription factors such as wrky, erf, mad, mikc, hsf and bzip were identified in mutant genotypes. Li et al. (2017) conducted comparative transcription analysis of kenaf under salt stress, identified 2 384 differentially expressed genes between salt stress and control plants, and identified 15 transcription factor families and 37 transcripts responding to salt stress. Zheng et al. (2020) conducted transcriptome analysis on maize inbred lines with different drought tolerance and revealed that the difference in drought tolerance of maize may be related to active oxygen scavenging capacity, signal interaction network and some transcription factors from the level of gene regulation. In this study, low-temperature treatment transcriptome sequencing was conducted for two eggplant materials with different low-temperature tolerance, and correlation analysis of differentially expressed genes obtained under low-temperature treatment and room temperature conditions could effectively help us understand the genetic characteristics of related materials at the molecular level, thus contributing to the acquisition and utilization of excellent low-temperature resistant materials.
3 Materials and Methods
3.1 Experimental materials
Two experimental materials were selected as the high generation eggplant inbred line material CHEN18 with strong cold tolerance and the high generation eggplant inbred line material 819 with high cold sensitivity, which were found by our research group in long-term eggplant breeding.
3.2 Selection of low temperature tolerance for seed germination of experimental materials
300 seeds of each material with high gloss and plumpness were taken and arranged neatly in the germination box with double-layer germination paper. Put the germination box in the biochemical incubator with the temperature of 5°C, 10°C, 15°C, 20°C, and 28°C respectively for seed germination test. Placed 100 seeds in each germination box, repeat for 3 times, and cleaned the surface of seeds with distilled water every day. Counted the seed germination every day, counted the germination rate of all seeds on the 14th day, and measured the germination index and vitality index according to the daily seed germination. Germination rate (Gr)=∑Nt/N×100%, Germination index (Gi)=∑(Nt/Dt), Vitality index (Vi)=Gi×S. Where, Nt represents the number of germinated seeds on day t, Dtrepresents the number of germination days corresponding to the number of germinated seeds on one day, N represents the total number of seeds involved in the germination test, S represents the height of seedlings at the end of germination test (expressed in cm of seedling height).
3.3 Selection of low temperature resistance of experimental materials at seedling stage
Two eggplant material seedlings were transplanted into 32-hole trays and placed in an artificial incubator for seedling growth under the conditions of 25°C~28°C temperature, 60%~70% humidity and 14 h/10 h light/dark cycle. When the seedlings grew to four real leaves, they were treated at 4°C for 0 h, 12 h, 24 h, 48 h, 72 h, 96 h, 120 h, 144 h and 168 h, respectively. The phenotypes of treated seedlings were statistically analyzed for chilling injury (Li et al., 2012), and the standards were as follows:
Grade 0: the plant grows normally without chilling injury symptoms.
Grade 1: Only a few leaves with slightly shrunk and wilted edges.
Grade 2: Half of the leaves withered and died due to chilling injury.
Grade 3: More than half of the leaves withered and died due to chilling injury.
Grade 4: All died or close to death due to chilling injury.
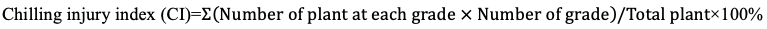
3.4 Low temperature treatment and sampling
According to seed germination and seedling low temperature test, the low temperature resistant material CHEN18 and cold sensitive material 819 of eggplant were tested. They were respectively seeded in 32-hole trays and placed in an artificial incubator for seedling growth under the temperature of 25°C~28°C, 60%~70% humidity and 14 h/10 h light/dark cycle. When the seedlings grow to four real leaves, they are treated at 4°C for 8 h. The leaves of the plants were sampled under the room temperature control condition and the low temperature treatment for 8 h and were quickly wrapped with tin foil and put into liquid nitrogen for freezing. After freezing, they were stored in the -80°C refrigerator. There were four treatments in the test: CHEN18 room temperature (CK1), CHEN18 low temperature 8 h (T1), 819 room temperature (CK2), 819 low temperature 8 h (T2). The sampling of each treatment shall be repeated for 3 times.
3.5 RNA Extraction, and construction and sequencing of cDNA Library
RNA was extracted with mirVana miRNA Isolation Kit (Ambion-1561) and purified with the RNeasy Plant Mini Kit (Qiagen). The concentration of total RNA was determined using the NanoDrop spectrophotometer of Thermo Fisher Scientific. The cDNA Library was constructed with the help of TruSeq Stranded mRNA LTSample Prep Kit from Illumina. After the total RNA was digested with DNase and enriched with magnetic beads with Oligo(dT), it was broken into small fragments of about 200 bp. And using this as a template, the first strand of cDNA was synthesized through SuperScript II Reverse Transcriptase (Invitrogen). The synthesis of the second strand depended on DNA polymerase I and RNase H. poly(A) tail was added to the end of double stranded cDNA and the sequencing connector was connected. The constructed cDNA Library was inspected with Agilent 2100 Bioanalyzer and sequenced with Illumina HiSeq X Ten sequencer after passing the inspection. Transcriptome sequencing was completed by Shanghai OE Biotech Co., Ltd.
3.6 RNA-seq analysis
HISAT2 software (Kim et al., 2015) was used to compare the clean reads obtained after removing the raw reads with connectors, high content of unknown base N and low-quality reads with the data in eggplant genome (http://eggplant.kazusa.or.jp/). The gene expression level is expressed by FPKM (the number of fragments per 1 000 base lengths of a transcript compared to one million fragments). The sequences of differentially expressed genes were compared and analyzed in KEGG (Kyoto Encyclopedia of Genes and Genomes) database with the help of BLASTX, and the protein with the highest similarity to the given sequence was retrieved and functional annotation was made. The GO (Gene Ontology) annotation of differentially expressed genes was obtained through Blast2GO, while WEGO (Web Gene Ontology Annotation Plot) software was used to classify GO functions. According to the analysis results, different differentially expressed genes were enriched into different GO entries, and the number of differentially expressed genes enriched in each entry was calculated. At the same time, the differentially expressed genes were analyzed with TBtools to perform Venn diagram.
Authors’ contributions
ZZW was the experimental designer and executor of this study and finished data sorting and the first draft of the paper. WXX and ZAD participated in parts of experiments. ZDS was the director of the project, guiding experimental design, data statistics, paper writing and revision. All authors read and approved the final manuscript.
Acknowledgments
This study was supported by the Youth Talents Development Plan of Shanghai Municipal Agricultural System (HNQZ (2018) No. 1-14) and the Fund for National Modern Agricultural Industrial Technology System Construction (CARS-23-G-40).
Bai B., Wu J., Zhang W., Yao D.P., Li Y.G., and Deng Q.Y., 2017, Studies on cold tolerance of widely adaptable photo-thermo-sensitive genic male sterile line Y58S and its physiological mechanism at booting stage, Zhiwu Yichuan Ziyuan Xuebao (Journal of Plant Genetic Resources), 18(4): 646-652
Bao C.L., Du L.M., Hu T.H., Zhu Q.M., Hu H.J., He Q.Y., and Mao W.H., 2013, Screening of low-temperature tolerance materials and studies on their physiological responses to low temperature in eggplant (Solanum melongena), Zhiwu Yichuan Ziyuan Xuebao (Journal of Plant Genetic Resources), 14(6): 1161-1166
Cheng J.Q., Shen D., Li X.X., Wang H.P., Qiu Y., and Song J.P., 2012, Field evaluation of low temperature tolerance of cucumber (Cucumis sativus) core collection, Zhiwu Yichuan Ziyuan Xuebao (Journal of Plant Genetic Resources), 13(4): 660-665
Deng R.J., Fan J.X., Wang Y.Q., Jin J.F., and Liu T., 2014, Physiological responses of Hylocereus undulatus Britt seedlings to low temperature stress and comprehensive evaluation of its cold resistance, Zhiwu Shengli Xuebao (Plant Physiology Journal), 50(10): 1529-1534
Dhawan S.S., Shukla P., Gupta P., and Lal R.K., 2016, A cold-tolerant evergreen interspecific hybrid of Ocimum kilimandscharicum and Ocimum basilicum: analyzing trichomes and molecular variations, Protoplasma, 253(3): 845-855
https://doi.org/10.1007/s00709-015-0847-9
PMid:26156173
Ebrahimi A., Zarei A., McKenna J.R., Bujdoso G., and Woeste K.E., 2017, Genetic diversity of Persian walnut (Juglans regia) in the cold-temperate zone of the United States and Europe, Scientia Horticulturae, 220: 36-41
https://doi.org/10.1016/j.scienta.2017.03.030
Huang Z., Zhang X.X., Jiang S.H., Qin M.F., Zhao N., Lang L.N., Liu Y.P., Tian Z.S., Liu X., Wang Y., Zhang B.B., and Xu A.X., 2017, Analysis of cold resistance and identification of SSR markers linked to cold resistance genes in Brassica rapa L, Breeding Science, 67(3): 213-220
https://doi.org/10.1270/jsbbs.16161
PMid:28744174 PMCid:PMC5515308
Kim D., Langmead B., and Salzberg S.L., 2015, HISAT: a fast spliced aligner with low memory requirements, Nature Methods, 12(4): 357-360
https://doi.org/10.1038/nmeth.3317
PMid:25751142 PMCid:PMC4655817
Li C., Tao R.F., Li Y., Duan M.H., and Xu J.H., 2020, Transcriptome analysis of the thermosensitive genic male-sterile line provides new insights into fertility alteration in rice (Oryza sativa). Genomics, 112(3): 2119-2129
https://doi.org/10.1016/j.ygeno.2019.12.006
PMid:31837402
Li H., Li D.F., Chen A.G., Tang H.J., Li J.J., and Huang S.Q., 2017, RNA-seq for comparative transcript profiling of kenaf under salinity stress, Journal of Plant Research, 130(2):365-372
https://doi.org/10.1007/s10265-016-0898-9
PMid:27999968 PMCid:PMC5318473
Li Y., Xie L.B., Chen Y.Q., and Fan S.Z., 2012, Effects of exogenous calcium chloride and abscisic acid on relative indexes of eggplant (Solanum melongena) under low temperature stress, Beifang Yuanyi (Northern Horticulture), (7): 22-25
Lian Y., Liu F.Z., Chen Y.H., 2006, Chinese eggplant cultivars (Solanum melongena L.) distribution and germplasm resources research advancement, Zhongguo Shucai (Chinese Vegetables), (10): 9-14
Lian Y., Liu F.Z., Tian S.B., Chen Y.H., and Zhang Y., 2017, Research progress on genetics and breeding of eggplant (Solanum melongena) in China during the 12th five-year plan, Zhongguo Shucai (Chinese Vegetables), (2): 14-22
Niño-Medina G., Urías-Orona V., Muy-Rangel M.D., and Heredia J.B., 2017, Structure and content of phenolics in eggplant (Solanum melongena)-a review, South African Journal of Botany, 111: 161-169
https://doi.org/10.1016/j.sajb.2017.03.016
Samaneh K.M., Reza M.A., and Seyyedeh-Sanam K.S., 2017, Effect of cold stress on oxidative damage and mitochondrial respiratory properties in chickpea, Plant physiology and biochemistry, 122: 31-39
https://doi.org/10.1016/j.plaphy.2017.11.011
PMid:29172103
Sareh Y., Hassan S., Seyedeh S.R., Khalil Z.N., and Vahid S., 2020, The RNA-seq transcriptomic analysis reveals genes mediating salt tolerance through rapid triggering of ion transporters in a mutant barley, PLoS one, 15(3): e0229513
https://doi.org/10.1371/journal.pone.0229513
PMid:32187229 PMCid:PMC7080263
Wang F., Chen X.X., Dong S.J., Jiang X.C., Wang L.Y., Yu J.Q., and Zhou Y.H., 2020, Crosstalk of PIF4 and DELLA Modulates CBF Transcript and Hormone Homeostasis in Cold Response in Tomato.Plant Biotechnol Journal, 18(4): 1041-1055
https://doi.org/10.1111/pbi.13272
PMid:31584235 PMCid:PMC7061876
Wang Y.X., Li W.S., Chang H., Zhou J.H., Luo Y.B., Zhang K.H., Zuo J.H., and Wang B.G., SRNAome and transcriptome analysis provide insight into strawberry fruit ripening, Genomics, 112(3): 2369-2378
https://doi.org/10.1016/j.ygeno.2020.01.008
PMid:31945464
Wu X.X., Zhu Z.W., Zhang AD., Xu S., Yao J., and Zha D.S., 2017, Effects of exogenous melatonin on growth, photosynthesis and antioxidant system of eggplant (Solanum melongena) seedlings under low temperature stress, Xibei Zhiwu Xuebao (Acta Bot. Boreal.-Occident. Sin), 37(12): 2427-2434
Wu Y.Y., Hao Y.C., Su Z.H., Guo S.J., Cao X.S., and Ji L.S., 2019, Screening of low temperature introgression lines and the differences in physiological response under cold stress of sweet pepper hardy germplasm, Fenzi Zhiwu Yuzhong (Molecular Plant Breeding), 17(4): 1249-1256
Yan L., Cai J.S., Gao L.B., Huang B., Ma H.Q., Liu Q.Y., Dai X.Y., Zhang X.K., Cheng Y., and Zou X.L., 2018, Establishment of cold resistance identification method and selection of germplasm resources in Brassica Napus L, Zhongguo Youliao Zuowu Xuebao (Chinese Journal of Oil Crop Sciencies), 40(1): 74-83
Zhang X.Y., and Xu K., 2009, Effect of interaction between rootstock and scion on cold resistance of grafted eggplant (Solanum melongena) seedlings under low temperature and low light, Zhongguo Nongye Kexue (Agricultural Science of China), 42(10): 3734-3740
Zhang Y.J., Su P., Qi J.B., Zhang Y., Zhang Y.T., Chen Y.B., Li Z.H., and Zhang Y.H., 2020, Effect of exogenous Nitric Oxide on resistance of eggplant (Solanum melongena) seedlings under low temperature stress, Zhiwu Shengli Xuebao (Plant Physiology Journal), 56(1): 66-72
Zhao R.L., Zhao Y., Xu K., Li J.H., Zhang S.H., and Yang X.J., 2019, Study on identification of wheat cold resistance by indoor freezing method, Zhiwu Yichuan Ziyuan Xuebao (Journal of Plant Genetic Resources), 20(2): 284-296
Zheng H.X., Yang Z., Wang W.Q., Guo S.J., LiZ. X., Liu K.C., and Sui N., 2020, Transcriptome analysis of maize inbred lines differing in drought tolerance provides novel insights into the molecular mechanisms of drought responses in roots, Plant Physiology and Biochemistry, 149: 11-26
https://doi.org/10.1016/j.plaphy.2020.01.027
PMid:32035249
Zhu Z.W., Wu X.X., Zhang A.D., Tian S.B., and Zha D.S., 2019, Effects of low temperature stress on seed germination of different eggplant (Solanum melongena) genotypes, Jiangxi Nongye Xuebao (Journal of Jiangxi Agricultural Science), 31(10): 39-44
. PDF(941KB)
. HTML
Associated material
. Readers' comments
Other articles by authors
. Zongwen Zhu
. Xuexia Wu
. Aidong Zhang
. Dingshi Zha
Related articles
. Solanum melongena L.
. Low temperature
. Transcriptome analysis
. Differentially expressed genes
Tools
. Email to a friend
. Post a comment
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)


